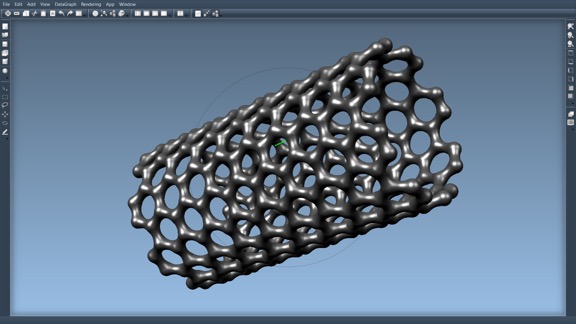
HyperBalls now in UnityMol!
04/08/2016 Rangé dans: News
The HyperBalls shaders are fully functional within Unity3D and are used in the UnityMol molecular viewer. HyperBalls are now fully integrated into the UnityMol software and are actively developed in that context. For instance they were extended with MatCap functionality to use lit spheres lighting. You will find the most up-to-date versions of the shaders there. They have been greatly optimized for use in a VR context for instance and run on a variety of devices such as Android tablets (Asus Nexus 7,..) and Android TVs, in a WebGL context, etc. Of course, they still run on MacOSX, Windows and Linux, too.
So if you are interested in HyperBalls, please follow the UnityMol project. There is a RSS feed, a twitter account and a Gitter chatroom available. The latest UnityMol build v 0.9.6 has been released today!
So if you are interested in HyperBalls, please follow the UnityMol project. There is a RSS feed, a twitter account and a Gitter chatroom available. The latest UnityMol build v 0.9.6 has been released today!
Hyperballs implemented in the NGLViewer
Hyperballs links are in the NGLviewer (http://arose.github.io/ngl/). Looks really good! Thanks @asrmoin !

New paper: Visualization of Molecular Structure: The State of the Art
11/12/2015 Rangé dans: Paper
Structural properties of molecules are of primary concern in many fields. This report provides a comprehensive overview on techniques that have been developed in the fields of molecular graphics and visualization with a focus on applications in structural biology. The field heavily relies on computerized geometric and visual representations of three-dimensional, complex, large, and time-varying molecular structures. The report presents a taxonomy that demonstrates which areas of molecular visualization have already been extensively investigated and where the field is currently heading. It discusses visualizations for molecular structures, strategies for efficient display regarding image quality and frame rate, covers different aspects of level of detail, and reviews visualizations illustrating the dynamic aspects of molecular simulation data. The report concludes with an outlook on promising and important research topics to enable further success in advancing the knowledge about interaction of molecular structures.
B. Kozlíková, M. Krone, N. Lindow, M. Falk, M. Baaden, D. Baum, I. Viola, J. Parulek, H.-C. Hege: Visualization of Molecular Structure: The State of the Art; Eurographics Conference on Visualization (EuroVis) (2015) edited by R. Borgo, F. Ganovelli, and I. Viola (Editors)
B. Kozlíková, M. Krone, N. Lindow, M. Falk, M. Baaden, D. Baum, I. Viola, J. Parulek, H.-C. Hege: Visualization of Molecular Structure: The State of the Art; Eurographics Conference on Visualization (EuroVis) (2015) edited by R. Borgo, F. Ganovelli, and I. Viola (Editors)
New paper: Three-Dimensional Representations of Complex Carbohydrates and Polysaccharides. SweetUnityMol: A Video Game Based Computer Graphic Software
01/12/2014 Rangé dans: Paper
A molecular visualization program tailored to deal with the range of 3D structures of complex carbohydrates and polysaccharides, either alone or in their interactions with other biomacromolecules, has been developed using advanced technologies elaborated by the video games industry. All the specific structural features displayed by the simplest to the most complex carbohydrate molecules have been considered and can be depicted. This concerns the monosaccharide identification and classification, conformations, location in single or multiple branched chains, depiction of secondary structural elements and the essential constituting elements in very complex structures. Particular attention was given to cope with the accepted nomenclature and pictorial representation used in glycoscience. This achievement provides a continuum between the most popular ways to depict the primary structures of complex carbohydrates to visualizing their 3D structures while giving the users many options to select the most appropriate modes of representations including new features such as those provided by the use of textures to depict some molecular properties. These developments are incorporated in a stand-alone viewer capable of displaying molecular structures, biomacromolecule surfaces and complex interactions of biomacromolecules, with powerful, artistic and illustrative rendering methods. They result in an open source software compatible with multiple platforms, i.e., Windows, MacOS and Linux operating systems, web pages, and producing publication-quality figures. The algorithms and visualization enhancements are demonstrated using a variety of carbohydrate molecules, from glycan determinants to glycoproteins and complex protein–carbohydrate interactions, as well as very complex mega-oligosaccharides and bacterial polysaccharides and multi-stranded polysaccharide architectures.
S. Pérez*, T. Tubiana, A. Imberty, M. Baaden: Three-Dimensional Representations of Complex Carbohydrates and Polysaccharides. SweetUnityMol: A Video Game Based Computer Graphic Software Glycobiology 25, 2015, 483-491
S. Pérez*, T. Tubiana, A. Imberty, M. Baaden: Three-Dimensional Representations of Complex Carbohydrates and Polysaccharides. SweetUnityMol: A Video Game Based Computer Graphic Software Glycobiology 25, 2015, 483-491
SAMSON features HyperBalls representations
24/03/2013 Rangé dans: News
The SAMSON Software for Adaptive Modelling and Simulation Of Nanosystems features HyperBalls visual representations. Here an early screenshot from the implementation phase.

New paper: Game on, Science - how video game technology may help biologists tackle visualization challenges
06/03/2013 Rangé dans: Paper
The video games industry develops ever more advanced technologies to improve rendering, image quality, ergonomics and user experience of their creations providing very simple to use tools to design new games. In the molecular sciences, only a small number of experts with specialized know-how are able to design interactive visualization applications, typically static computer programs that cannot easily be modified. Are there lessons to be learned from video games? Could their technology help us explore new molecular graphics ideas and render graphics developments accessible to non-specialists? This approach points to an extension of open computer programs, not only providing access to the source code, but also delivering an easily modifiable and extensible scientific research tool. In this work, we will explore these questions using the Unity3D game engine to develop and prototype a biological network and molecular visualization application for subsequent use in research or education. We have compared several routines to represent spheres and links between them, using either built-in Unity3D features or our own implementation. These developments resulted in a stand-alone viewer capable of displaying molecular structures, surfaces, animated electrostatic field lines and biological networks with powerful, artistic and illustrative rendering methods. We consider this work as a proof of principle demonstrating that the functionalities of classical viewers and more advanced novel features could be implemented in substantially less time and with less development effort. Our prototype is easily modifiable and extensible and may serve others as starting point and platform for their developments. A webserver example, standalone versions for MacOS X, Linux and Windows, source code, screen shots, videos and documentation are available at the address: http://unitymol.sourceforge.net/.
Z. Lv, A. Tek, F. Da Silva, C. Empereur-mot, M. Chavent and M. Baaden: Game on, Science - how video game technology may help biologists tackle visualization challenges PLoS ONE 8(3):e57990.
Z. Lv, A. Tek, F. Da Silva, C. Empereur-mot, M. Chavent and M. Baaden: Game on, Science - how video game technology may help biologists tackle visualization challenges PLoS ONE 8(3):e57990.
Version controlled source code available via svn
16/10/2011 Rangé dans: Code
We have set up a subversion repository for the demo source code. It now works cross-platform (MacOSX, Linux, Windows) and we have added a few command line options as well as SpaceBall and Joystick support. The Tutorial subpage explains in detail how to install this code using CMake. Current version is v0.2, and we are still in the process of refining and cleaning up the code.
GLSL version available
10/09/2011 Rangé dans: Release
A GLSL version of the simple HyperBalls demo can now be downloaded from the Sourceforge website (http://sourceforge.net/projects/hyperballs/files/HyperBallDemo_V0.1_glsl.tgz/download). The GLSL version is compatible with a wide variety of graphics cards. Still, the graphics card must support at least Shader Model 3.0.
Journal of Computational Chemistry: GPU-accelerated atom and dynamic bond visualization using hyperballs: A unified algorithm for balls, sticks, and hyperboloids
17/08/2011 Rangé dans: Paper
 The main HyperBalls article is now available at the Journal of Computational Chemistry. The title is "GPU-accelerated atom and dynamic bond visualization using hyperballs: A unified algorithm for balls, sticks, and hyperboloids".
The main HyperBalls article is now available at the Journal of Computational Chemistry. The title is "GPU-accelerated atom and dynamic bond visualization using hyperballs: A unified algorithm for balls, sticks, and hyperboloids".Keywords: GPU computing;ray-casting;improved ball-and-stick and licorice representations;HyperBalls;pixel accurate representation
Abstract: Ray casting on graphics processing units (GPUs) opens new possibilities for molecular visualization. We describe the implementation and calculation of diverse molecular representations such as licorice, ball-and-stick, space-filling van der Waals spheres, and approximated solvent-accessible surfaces using GPUs. We introduce HyperBalls, an improved ball-and-stick representation replacing tubes, linking the atom spheres by hyperboloids that can smoothly connect them. This type of depiction is particularly useful to represent dynamic phenomena, such as the evolution of noncovalent bonds. It is furthermore well suited to represent coarse-grained models and spring networks. All these representations can be defined by a single general algebraic equation that is adapted for the ray-casting technique and is well suited for execution on the GPU. Using GPU capabilities, this implementation can routinely, accurately, and interactively render molecules ranging from a few atoms up to huge macromolecular assemblies with more than 500,000 particles. In simple cases, based only on spheres, we have been able to display up to two million atoms smoothly.
By: Matthieu Chavent, Antoine Vanel, Alex Tek, Bruno Levy, Sophie Robert, Bruno Raffin, Marc Baaden
Journal of Computational Chemistry, Volume 32, Issue 13, pages 2924–2935, October 2011
HyperBalls demo source code now online !
17/06/2011 Rangé dans: Code
It now is possible to download the code of a simple HyperBalls demo and compile it (http://sourceforge.net/projects/hyperballs/files/HyperBallDemo_V0.1_Cg.tgz/download).
There are still a few constraints, first related to availability. For the moment, the code works quite well on a Linux machine, but not on MacOs X. This is due to a limitation of the current OSX OpenGL implementation and we are working on alternatives. We will soon try to test the demo on a Windows machine.
Second, the hardware requirements: The current version of the program only works on Nvidia cards (Cg shaders). We will very soon provide a GLSL version, compatible with a wide variety of graphics cards. The graphics card must support at least Shader Model 3.0.
We hope to provide more demos and code soon, so stay tuned and send us your feedback!
There are still a few constraints, first related to availability. For the moment, the code works quite well on a Linux machine, but not on MacOs X. This is due to a limitation of the current OSX OpenGL implementation and we are working on alternatives. We will soon try to test the demo on a Windows machine.
Second, the hardware requirements: The current version of the program only works on Nvidia cards (Cg shaders). We will very soon provide a GLSL version, compatible with a wide variety of graphics cards. The graphics card must support at least Shader Model 3.0.
We hope to provide more demos and code soon, so stay tuned and send us your feedback!