Performance:
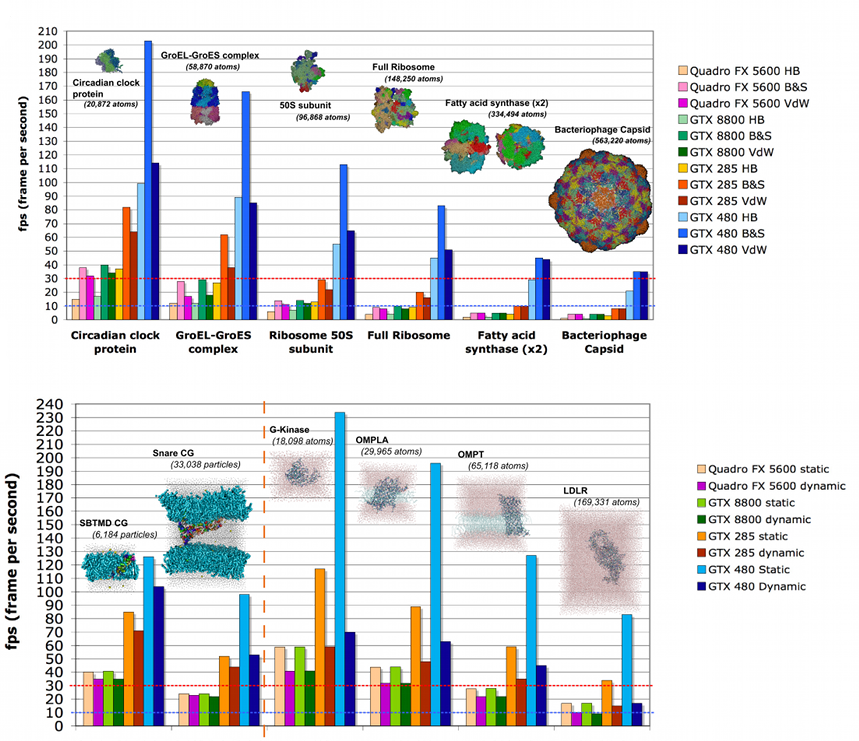
For the static benchmark, we used a dataset with macromolecules ranging from approximately 20,000 to more than 550,000 atoms (see figure below). Each dataset was displayed using three different types of representations: HyperBalls (HB), ball & stick (B&S) and van der Waals (VdW). The benchmark measures the screen refresh rate in frames per second (fps).
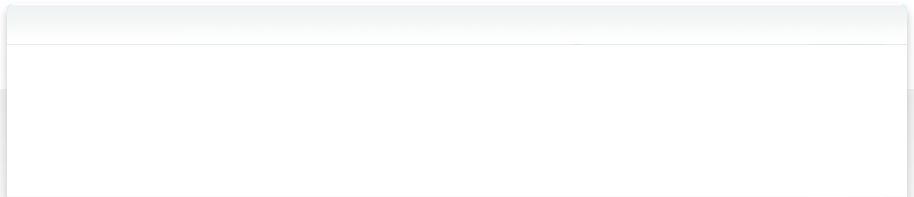
We have also assessed the visualization performance on a range of multiple time step systems. To benchmark evolving data originating from molecular dynamics simulations, we have created a simple trajectory viewer that reads Gromacs trajectory files and visualizes them.
At present, without further optimization, we can smoothly and interactively render systems with more than 550,000 atoms, reaching some limits for systems comprising a few million spheres. An encouraging feature is that we observe a significant enhancement of the efficiency of the algorithm for each new generation of graphics cards. It can be expected that this limit will be overcome with the next generation of graphics cards.
More information in the HyperBalls paper (see citation section for the paper reference).
Warning:
The values presented in this figure were taken from the HyperBalls paper and were obtained using the FvNano framework. Fps values can change a little bit with respect to the HyperBalls demo program provided on this website, as the code was kept simpler in order to maintain a good readability.