HyperBallView demo v0.2 README (under construction)
$Id: README.txt 10 2011-10-16 15:14:20Z baaden $
HyperBallView is a demo program illustrating the HyperBalls representation (Chavent et al., 2011) designed to visualize molecular structures using GPU graphics card capabilities based on shaders (GLSL or Cg). It can read Protein Data Bank (PDB) files and a custom netCDF format.
The code is also intended as an example that can be re-used to implement the HyperBalls representation in other software. The performance of the code is not maximal, as this would require restructuring the program and it would become less readable.
More information on http://hyperballs.sourceforge.net
Features
- fast HyperBalls (Chavent et al., 2011) shaders for continuous representations including balls, sticks and intermediates
- interactively control HyperBalls parameters such as shrink factor and atom/bond radii
- navigate the sceen with a SpaceBall (for example the SpaceNavigator) or a joystick via GLUT
- choose among several color schemes; define background color
- read PDB files and netCDF files from the FvNano project
- cross-platform application tested on MacOSX, Linux and Windows with shaders in GLSL and Cg languages
Installation
Dependencies
The demo uses CMake to facilitate cross-platform installations. Several graphics libraries are also needed.
Required
First, install the following dependencies: CMake, Cg Toolkit, freeglut/glut and a standard development environment (compiler and header files).
Optional
To install from the very latest source code version (recommended) you need the subversion svn utility. If you want to use the netCDF input files, you will also need the netCDF library and header files.
Compilation
Download source code
We advise you to get the latest version from svn, and put it in the ${HBDEMO_SRC} directory of your liking. In the next parts, we will refer to the absolute path of this directory as HBDEMO_SRC. So please replace it by your own path: for example it could be defined as /home/martin/hyperballdemo via the following command
export HBDEMO_SRC=/home/martin/hyperballdemo (for bash)or
setenv HBDEMO_SRC /home/martin/hyperballdemo (for csh)Now you can check out the latest version from the subversion repository using
svn co https://hyperballs.svn.sourceforge.net/svnroot/hyperballs ${HBDEMO_SRC}This creates the demo source directory.
Compile and install the demo
mkdir HBDemoBuild cd HBDemoBuild cmake ${HBDEMO_SRC} make install (you may have to be root)This will install the demo in the default directory determined by the install prefix (/usr/local, be careful you might need to be root). You can also install the demo on your own install prefix (for example /home/martin) using this way :
mkdir HBDemoBuild cd HBDemoBuild cmake ${HBDEMO_SRC} -DCMAKE_INSTALL_PREFIX:PATH=/home/martin make installLet's call the install prefix ${HBDEMO_EXE}. The above commands should generate and install an executable shell script called HBView_demo in ${HBDEMO_EXE}/bin.
Usage
The current demo is intentionally kept simple and only a subset of functionalities is available. For the moment you can load a molecule and change some visualization parameters. The atom color is fixed at startup. As the code is very simple, you can easily tune more colors and so on. The main goal of this demo is to show how to use the shaders (code for the GPU) to display molecular representations.
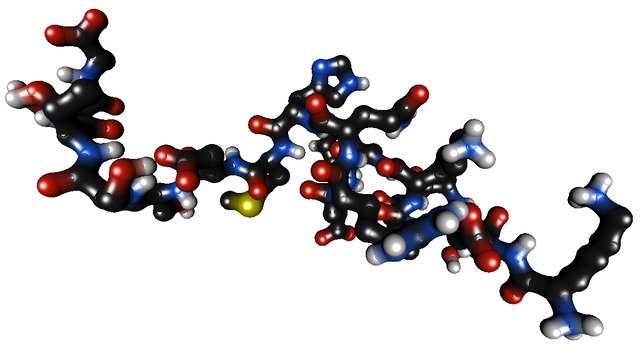
To launch the viewer, use a pdb file. There is a molecule test provided in the file example.pdb. To visualize it, do:
${HBDEMO_EXE}/bin/HBView_demo ${HBDEMO_SRC}/share/example.pdb
This should yield an image similar to the one below (eventually with different colors).
Command line options
- -help: Prints a short help text
- -bcol (int): Change background color: 0=white (default), 1=black, 2=grey
- -acol (int): Change atom color scheme: 0=CPK (default), 1=VMD-like, 3=test,...
- -noinfo: Disable on-screen information about radii etc.
Change the visual representation
You can interactively change the shrink factor (s key):
- increase shrink: S
- diminish shrink: s
You can interactively change the atom radii (a key):
- increase atom radii: A
- diminish atom radii: a
You can interactively change the bond radii (b key):
- increase bond radii: B
- diminish bond radii: b
You can see the values of these parameters on the screen (unless you have chosen the -noinfo command line option).
Just push the esc key to quit the demo.
You can also switch between standard representations (number keys):
- Ball & Stick rep. : 1
- Licorice rep. : 2
- VdW rep. : 3
- HyperBalls rep. (smooth links): 4
Navigate in the scene
Mouse and keyboard
Rotation/translation:
- You can rotate the molecule with the mouse clicking on the left button.
- You can translate the molecule with the mouse clicking on the right button.
- You can also translate the molecule using arrow (cursor) keys.
Zoom in/Zoom out (z key):
- You can zoom in with z
- You can zoom out with Z.
- You can zoom the molecule with the mouse clicking on the central button.
SpaceBall
We tested the 3DConnexion SpaceNavigator on MacOSX and configured it such that you can simultaneously rotate and translate the view along all three axes. The axis mapping may be machine-specific, so you may have to modify it. Please let us know if this is the case.
Joystick
We tested the Logitech Rumblepad on MacOSX for navigation in the scene. Button and stick mapping is still being improved. Any suggestions and contributions are welcome.
Citation
- M. Chavent, A. Vanel, A. Tek, B. Levy, S. Robert, B. Raffin, M. Baaden: GPU-accelerated atom and dynamic bond visualization using HyperBalls, a unified algorithm for balls, sticks and hyperboloids; J Comput Chem (2011) 32, 2924.
Related work
- M. Chavent, B. Levy, M. Krone, K. Bidmon, J. P. Nomine, T. Ertl and M. Baaden: GPU-powered tools boost molecular visualization; Brief Bioinformatics (2011)
Troubleshooting
- please note that command line parsing is very basic. Wrong number/type of arguments will crash the software
The end!
We hope you enjoy interactive HyperBalls visualization via this demo. Please let us know about your applications and discoveries!
Copyright and License
This program is under the CeCill licence, which is compatible with the GPL licence.
Appendix
Appendix 1: complementary installation instructions
To be completed soon for Windows and Linux.
MacOSX specific
- if you want to use Xcode, use cmake like this: cmake -G Xcode ${HBDEMO_SRC}
- if you get an error "CMake Error: The following variables are used in this project, but they are set to NOTFOUND." then you need to define some values on the cmake command line, In particular:
- for "GLUT_INCLUDE_DIR (ADVANCED)" add something like "-DGLUT_INCLUDE_DIR=/Developer/SDKs/MacOSX10.6.sdk/System/Library/Frameworks/GLUT.framework/Versions/A/Headers/"
- for "OPENGL_INCLUDE_DIR (ADVANCED)" add something like "-DOPENGL_INCLUDE_DIR=/Developer/SDKs/MacOSX10.6.sdk/System/Library/Frameworks/OpenGL.framework/Versions/A/Headers/"
- For example, in my case, the full line looks like this:
- cmake -G Xcode ${HBDEMO_SRC} -DCMAKE_INSTALL_PREFIX:PATH=/scratch/hbexe -DGLEW_LIBRARY=/opt/src/HyperBalls/glew151/x86_64/libGLEW.a -DGLUT_INCLUDE_DIR=/Developer/SDKs/MacOSX10.6.sdk/System/Library/Frameworks/GLUT.framework/Versions/A/Headers/ -DOPENGL_INCLUDE_DIR=/Developer/SDKs/MacOSX10.6.sdk/System/Library/Frameworks/OpenGL.framework/Versions/A/Headers/