News & Blog
Latest news updates available | Blog posts below

Latest news updates available | Blog posts below

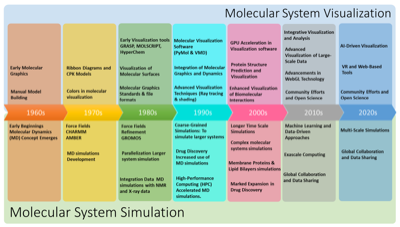 Recently, I had the opportunity to contribute to two review articles on the visualization of molecular dynamics simulations. The first one deals with this topic in depth, while the second article takes a more targeted approach and focuses on membrane systems in a concise perspective. The first review article addresses challenges and opportunities in visualizing complex MD simulations. It emphasizes the need for novel visual representations tailored to the dynamics and intricacies of large biomolecular systems. In our work, we present a classification scheme based on a visual abstraction formalism that serves as a guiding framework and identifies potential areas for future progress. In the second manuscript, we address the current challenge of visualizing the complex dynamics of membrane systems. We provide a historical overview of the development of visualization techniques and trace their evolution from simple line representations in the 1980s to sophisticated graphics and virtual reality applications today.
Recently, I had the opportunity to contribute to two review articles on the visualization of molecular dynamics simulations. The first one deals with this topic in depth, while the second article takes a more targeted approach and focuses on membrane systems in a concise perspective. The first review article addresses challenges and opportunities in visualizing complex MD simulations. It emphasizes the need for novel visual representations tailored to the dynamics and intricacies of large biomolecular systems. In our work, we present a classification scheme based on a visual abstraction formalism that serves as a guiding framework and identifies potential areas for future progress. In the second manuscript, we address the current challenge of visualizing the complex dynamics of membrane systems. We provide a historical overview of the development of visualization techniques and trace their evolution from simple line representations in the 1980s to sophisticated graphics and virtual reality applications today. Lately, it has been quite silent here on my blog. I have been moving my whole workflow and toolchain to a new paradigm, so I put the production of new content on hold until I started to get a working implementation. This is now slowly coming together, but it is still not quite there yet. For now I will minimally add new information here, and later on transition all the current content to the new system. Here are a few brief snippets of information.
Lately, it has been quite silent here on my blog. I have been moving my whole workflow and toolchain to a new paradigm, so I put the production of new content on hold until I started to get a working implementation. This is now slowly coming together, but it is still not quite there yet. For now I will minimally add new information here, and later on transition all the current content to the new system. Here are a few brief snippets of information. Recently, my colleagues and I published a scientific paper in the journal Algorithms on an algorithm that allows for fast and interactive positioning of proteins within membranes. The original model was strongly inspired by Brasseur's work from the end of the 90s.
Recently, my colleagues and I published a scientific paper in the journal Algorithms on an algorithm that allows for fast and interactive positioning of proteins within membranes. The original model was strongly inspired by Brasseur's work from the end of the 90s.