Tutorial and Guidance
QuickStart
version 1.0.25beta
UnityMol User Interface
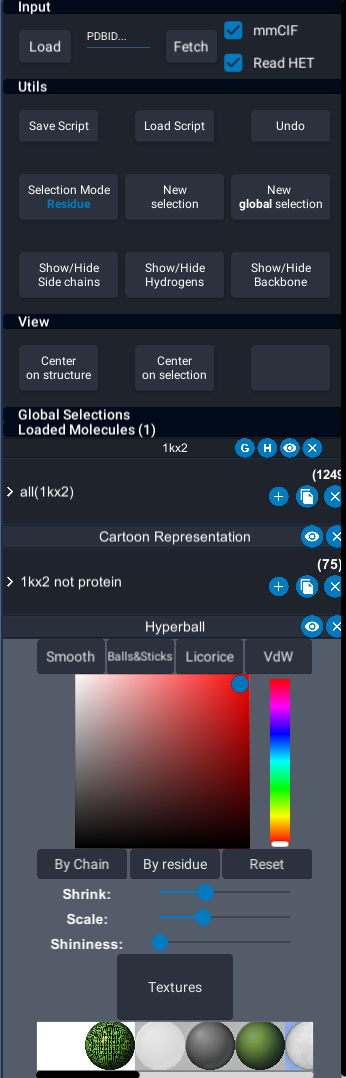
This UI was proposed to provide all UnityMol features in an accessible way.
Structure label buttons
Each loaded molecule has a label with button : 
- The 1st button (G) is to define groups to move them together in VR.
- The 2nd (H) is a utility tool to call Reduce (http://kinemage.biochem.duke.edu/software/reduce.php) to add hydrogens to the molecule. This can be slow, the best option is to manage the PDB file outside of UnityMol.
- The 3rd button hides or shows every representations of the corresponding molecule.
- The 4th is to delete/unload the molecule
Selections
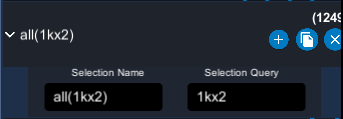
The selection button can be unfolded to show two input fields :
- The first one is the name of the selection.
- The second one can be used to enter a selection query using the selection language (see Selection language)
The (+) button allows to add representations to the corresponding selection. The (x) button deletes the selection and all the representations of this selection.
You can set or unset the current selection by clicking on the selection button, it will make the button darker and will show an outline around atoms of the selection. This outline is not updated when reading a trajectory for performance reasons for now.
Representations
First, the eye button hides/shows the corresponding representation. You can also delete a representation using the (x) button.
Note that if you modify the selection, you have to click on the eye button to update the representation.
By clicking on the selection label, the menu unfolds and you can tweak the different parameters of the representation.
Python console
Python integration in UnityMol is done using IronPython. It is a powerful python integration without needing to do every bindings for a function meaning that you can call every C# and Unity class/methods (for example GameObject.Find("Main Camera").transform.position = Vector3(0.1,0.2,0.3)).
However, not every python modules are available (eg. numpy, scipy, ...).
The console is based on https://github.com/AlexLemminG/PythonToUnity_Integration.
An API is defined in APIPython.cs and all the actions that you do from the UI do and should do a call to these API functions.
To show the console press the '!' key or '²' for qwerty keyboards
Some examples of commands in the APIPython
python
load("filePath")
python
fetch("1KX2")
python
select("1KX2 and resname CYS", "mySelection")
python
show("cartoon")
python
hide("hb")
python
showSelection("mySelection", "line")
python
hideSelection("mySelection", "line")
python
centerOnSelection("mySelection")
python
loadTraj("structureName", "filePath.xtc")
python
delete("structureName")
python
colorSelection("mySelection", "line", "red")
python
colorByChain("mySelection", "s")
python
renameSelection("mySelection", "myNewSelection")
python
screenshot("C:/Users/myUserName/Desktop/UMolScreenshot1.png")
python
startVideo("C:/Users/myUserName/Desktop/UMolVid1.mp4")
stopVideo()
python
connectIMD("myStructure", "127.0.0.1", 8888)
disconnectIMD("myStructure")
Selection language
Please refer to MDAnalysis documentation (here) for a detailed explanation about the language and some examples.
Select from the python console
python
select("water within 3.0 not protein") #default name of the selection will be "selection"
select("chain A and resid 1:10","name of my selection")
Note that select applies to all loaded molecules. If you want to apply only on a molecule do:
python
select("1KX2 and not protein")
Note that one can name a selection and use it in another selection, example:
```python select("1KX2 and ligand", "myLigandSel")
Record the myLigand keyword associated to myLigandSel selection
addSelectionKeyword("myLigand","myLigandSel")
select("myLigand and type FE","FELigand")
```
Examples of selections
python
select("prop y > 0.0", "aboveZero")
VR commands
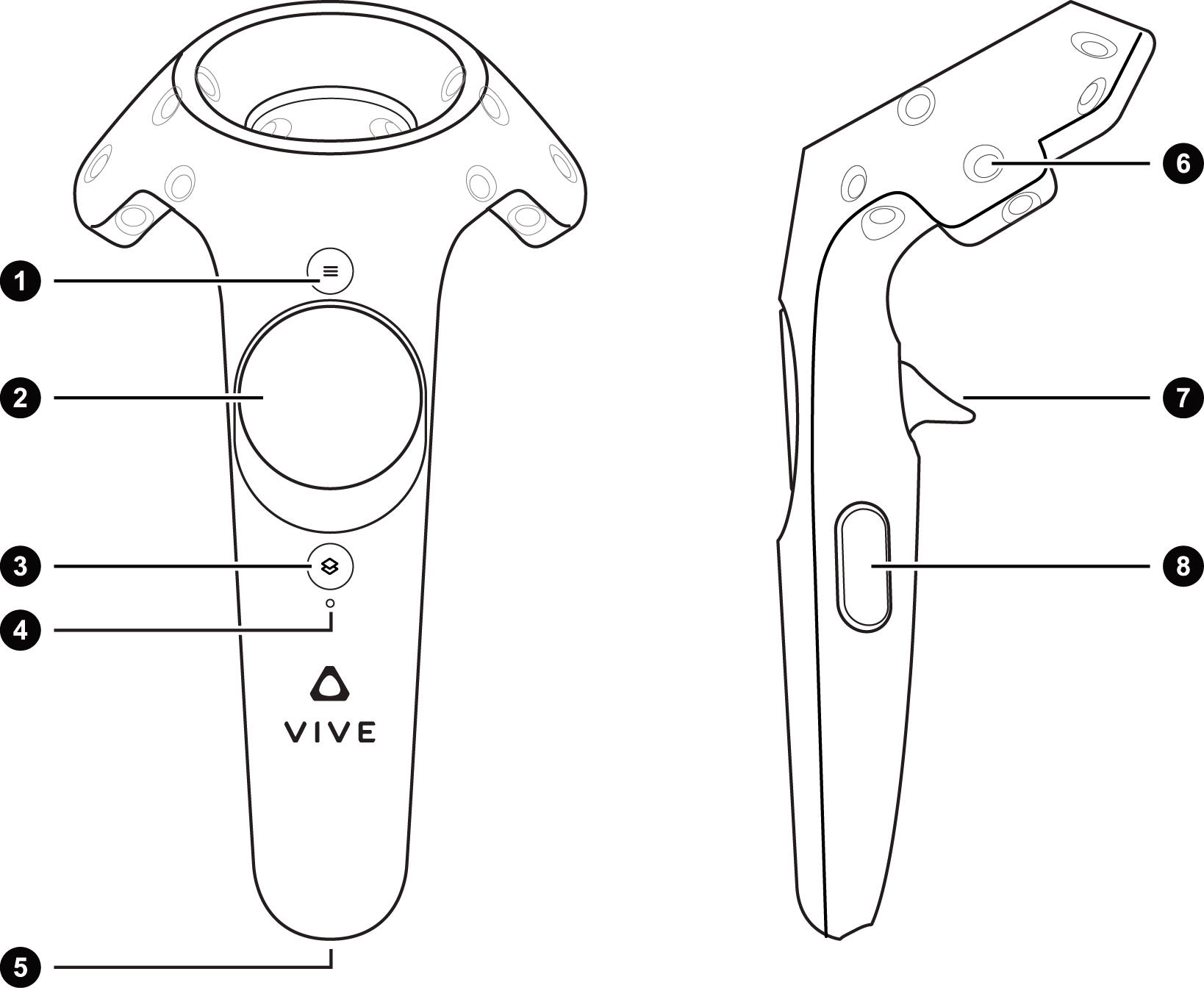
1- Measure distances, angles and torsion angles
2- Select/Deselect Residues, Chains or individual atoms| UI interaction
7- Move molecules around by pointing at them | Resize molecules using both controllers
- 8- Move all molecules of the same group
Trajectory reading
First load a molecule then load a trajectory file using the file browser or the command line. The trajectory is linked to the last loaded molecule.
UnityMol State file / Command history
UnityMol provides a way to save the state of the scene by saving all the executed commands in a text file of the form of a python script with UnityMol APIPython commands.
One can also create a script by hand a load it using the "Load script" button.
The script generated by the "Save script" button can contain a lot of redundant calls. It also contains the position of the loaded molecules parent.
Community
Unitymol is on twitter and we also have a Gitter community room. In addition there are three mailing lists visible through the forum pages, and a yet pristine support ticketing system.
(Older version < 1.0.0) Available resources
Check the movie for a first overview of UnityMol features. Several tutorials have been added to the user documentation available online. A few walkthroughs for older versions of UnityMol are available from the YouTube Channel.
You can still download a little guide here, but it is now largely outdated. A brief first steps user manual was put together here.
