Computational study of the outer membrane enzyme Ompla
We recently published computational work on OmPLA where we describe molecular dynamics simulations of a monomeric and of two dimeric OmPLA systems embedded in a lipid bilayer. [[Baaden 2003]] The above short movie (58 s, 11 MB) in DivX format illustrates OmPLA active site motions.
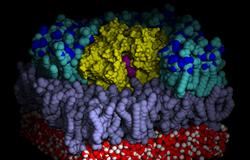
The above image shows the position of the HDS substrate in the binding pocket of the OmPLA dimer. Outer and inner leaflet of the bilayer are colored differently, and part of the upper layer as well as all extracellular water are omitted for clarity.
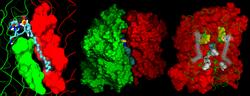
Above, the position of the HDS substrate at the OmPLA monomer-monomer interface is shown in a closeup view (left), the overall monomer-substrate-monomer system is illustrated (center) and the key interactions at the dimer interface are highlighted (right).
A collection of slides about our work on OmPLA can be accessed either online (OmPLA presentation foils), or downloaded as PDF file (OmPLA slides in PDF format).
The main tools used for the preparation of the above materials were VMD, Yasara, MSMS, Tachyon, transcode, iMovie, Raster3D and MagicPoint. © 2003-2005 by Marc Baaden.