FonFlon: “Relation fonction/fluctuation chez les proteines”. About the link between biological function and fluctuations for proteins.
Mosconi, Allemand, Bensimon, Delalande, Sacquin & Baaden 2007.
Latest News (updated 20/9/2011)
- A second paper investigating important states of guanylate kinase appeared in Biophysical journal (reference below)
- Our first paper on modeling tension-modulated enzymatic activity of guanylate kinase appeared in Biophysical journal (reference below).
- Press release (in French) concerning the DNase work.
- Second paper on the DNase I enzymatic mechanism published in PLOS Computational Biology (reference below).
- First paper on interactive exploration of cation binding to DNase I published in Pacific Symposium for Biocomputing (reference below).
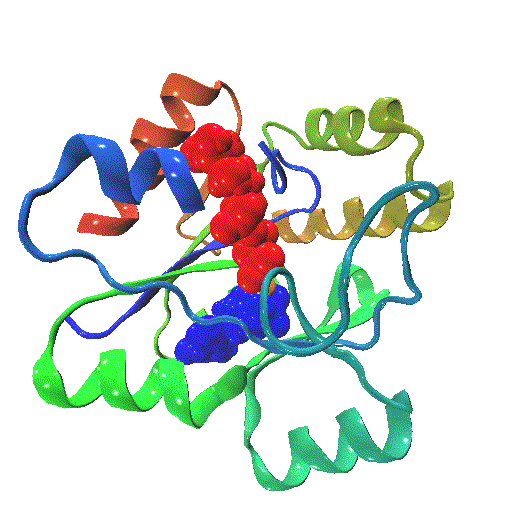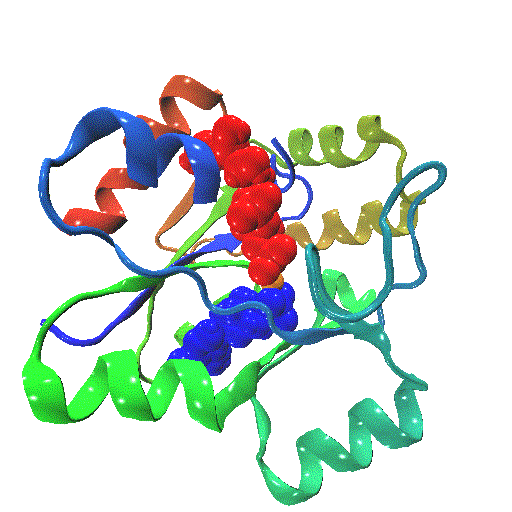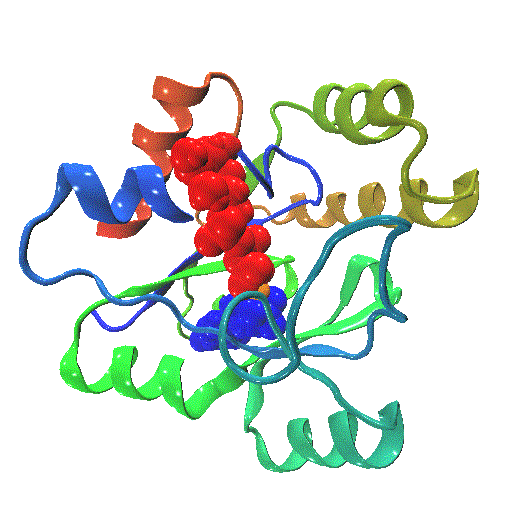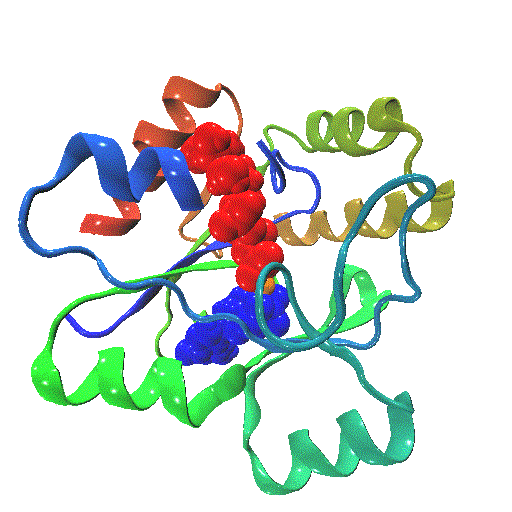
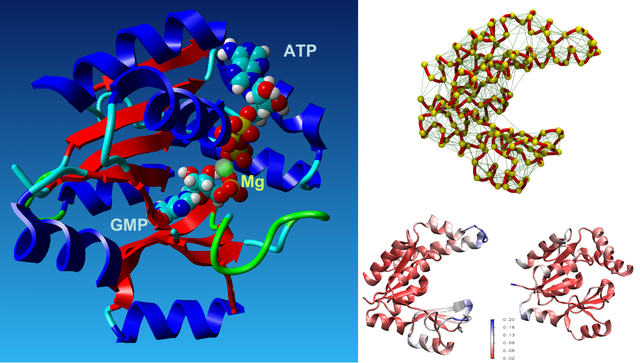
The closure mechanism of M. tuberculosis guanylate kinase relates structural fluctuations to enzymatic function
The Allosteric Spring Probe (ASP) technique allowed Zocchi et al. to act on the enzymatic activity of guanylate kinase (GK) by applying tension upon the molecular structure of this enzyme [Choi et al., Biophys. J., 2007]. These experiments raise the question about the underlying conformational modifications leading to such an observation.
In order to elucidate the results from these ASP studies, we investigate the conformational dynamics of GK and its mechanical properties. We use both high resolution atomistic molecular dynamics and low resolution Brownian Dynamics simulations. The enzyme is subject to large conformational changes, leading from an open to a closed form, and further influenced by substrate or co-factor docking. A reduction or perturbation of the conformational space available to GK can be related to the activity loss encountered in the ASP experiments.
We particularly focus on GK's closure mechanism with the aim to establish a detailed picture of the hierarchy and chronology of structural events essential for the enzymatic reaction. Rigidity profiles obtained from simulations of distinct states of the system hint at important differences. We have investigated open vs. closed, apo vs. holo or substrate vs. product-loaded forms of the enzyme.
Related Publications
- O. Delalande, S. Sacquin-Mora and M. Baaden : "Enzyme Closure and Nucleotide Binding Structurally Lock Guanylate Kinase", Biophysical Journal, 2011, 101, 1440
- M. Guéroult, D. Picot, J. Abi-Ghanem, B. Hartmann and M. Baaden : "How cations can assist DNase I in DNA binding and hydrolysis", PLOS Comput. Biol. 6, 2010, e1001000.
- S. Sacquin-Mora, O. Delalande and M. Baaden : "Functional modes and residue flexibility control the anisotropic response of Guanylate Kinase to mechanical stress", Biophys.J. 99, 2010, 3412-3419.
- O. Delalande, N. Férey, B. Laurent, M. Guéroult, B. Hartmann and M. Baaden: "Multi-resolution and multi-physics approach for interactively locating functionally linked ion binding sites by steering small molecules into electrostatic potential maps using a haptic device", Pac. Symp. Biocomput., 2010, 205-15.
- O. Delalande, N. Férey, G. Grasseau and M. Baaden : "Complex Molecular Assemblies at hand via Interactive Simulations", Journal of Computational Chemistry 30, 2009, 2375-2387
- N. Férey, O. Delalande, G. Grasseau and M. Baaden : "A VR framework for interacting with molecular simulations", 2008, in Proceedings of the 2008 ACM symposium on Virtual reality software and technology, edited by E. Kruiff, pp. 91-94, ACM, Bordeaux, France.
- N. Férey, O. Delalande, G. Grasseau and M. Baaden : "From Interactive to Immersive Molecular Dynamics", 2008, in Proceedings of the International Workshop on Virtual Reality and Physical Simulation (VRIPHYS'08), edited by F. Faure and M. Teschner, pp. 89-96, Eurographics, Grenoble, France.
You can also access my complete list of publications with links to the download locations.
Poster selection
Support
This project is supported by the ANR funding agency as project No ANR-06-PCVI-0025-01 in the PCV (Physique et Chimie du Vivant) program 2006.
Copyright
The FonFlon logo is copyright 2007 by Marc Baaden. It is licensed under the creative commons license (see page footer for details).